TogoTable retrieves annotations from distributed biological databases.
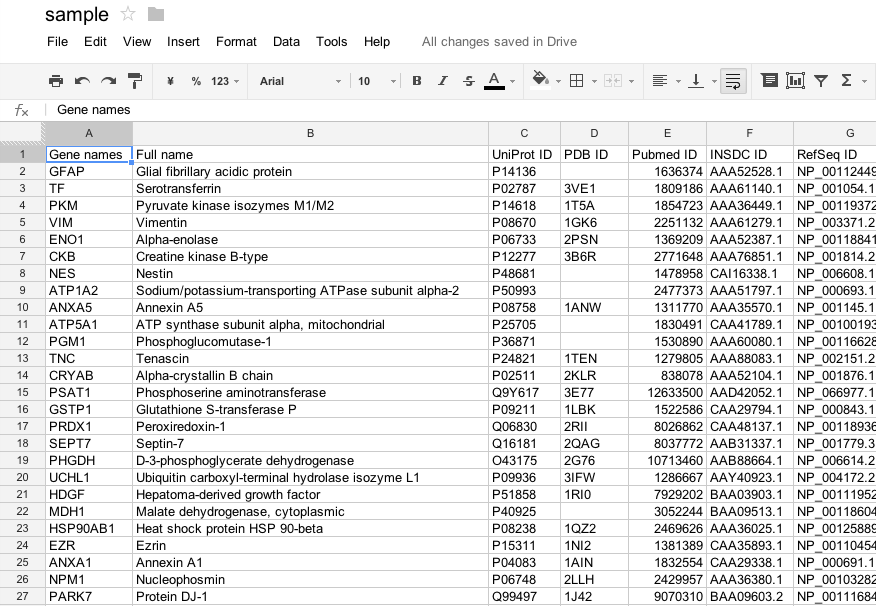
Prepare a tab separated value (TSV) file
Prepare a tabular data that contains biological IDs
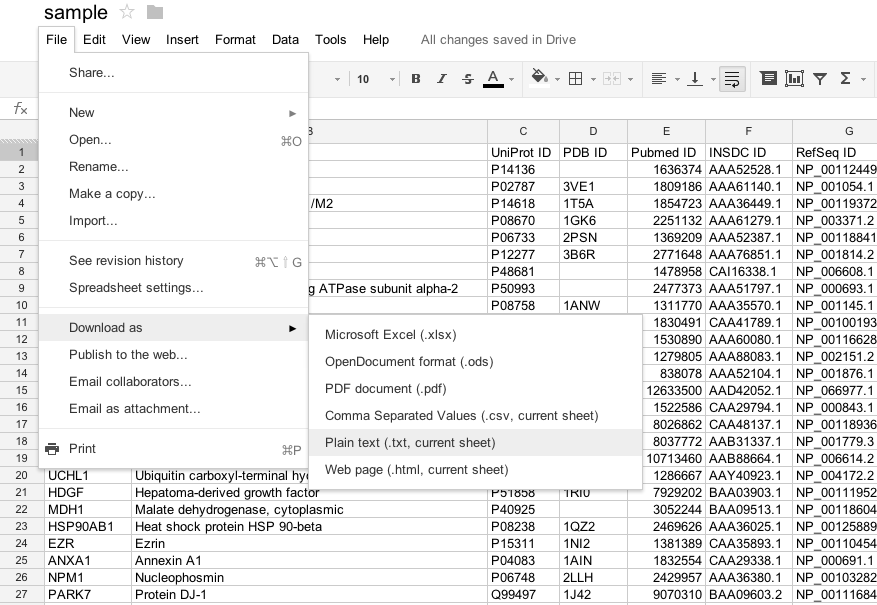
Save or export as the tab separated value (TSV) file
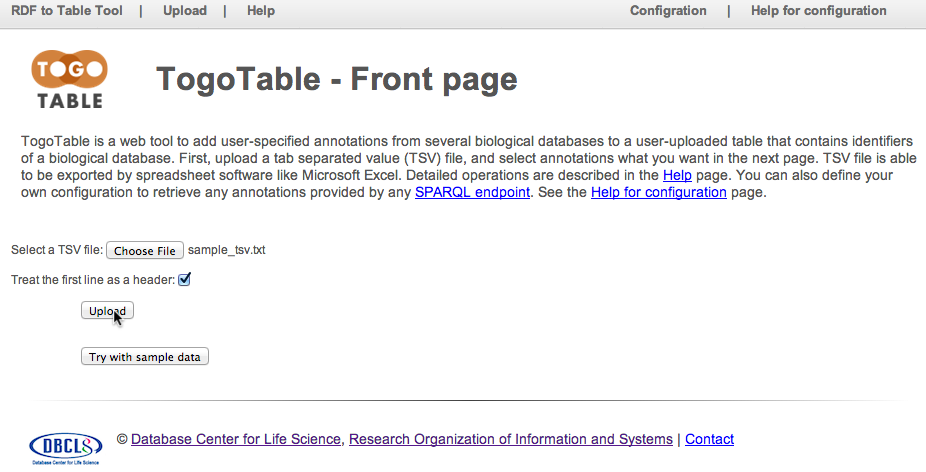
Select TSV file (or click "Try with sample data" in order to test the TogoTable service using an example) at the TogoTable website, specify whether the first line is header or not, and click "Upload".
Note that TogoTable only accepts a tab-separated text file. It cannot accept text files, whose delimiter is other than tab (e.g. space, comma, colon, and semi-colon), and binary files.
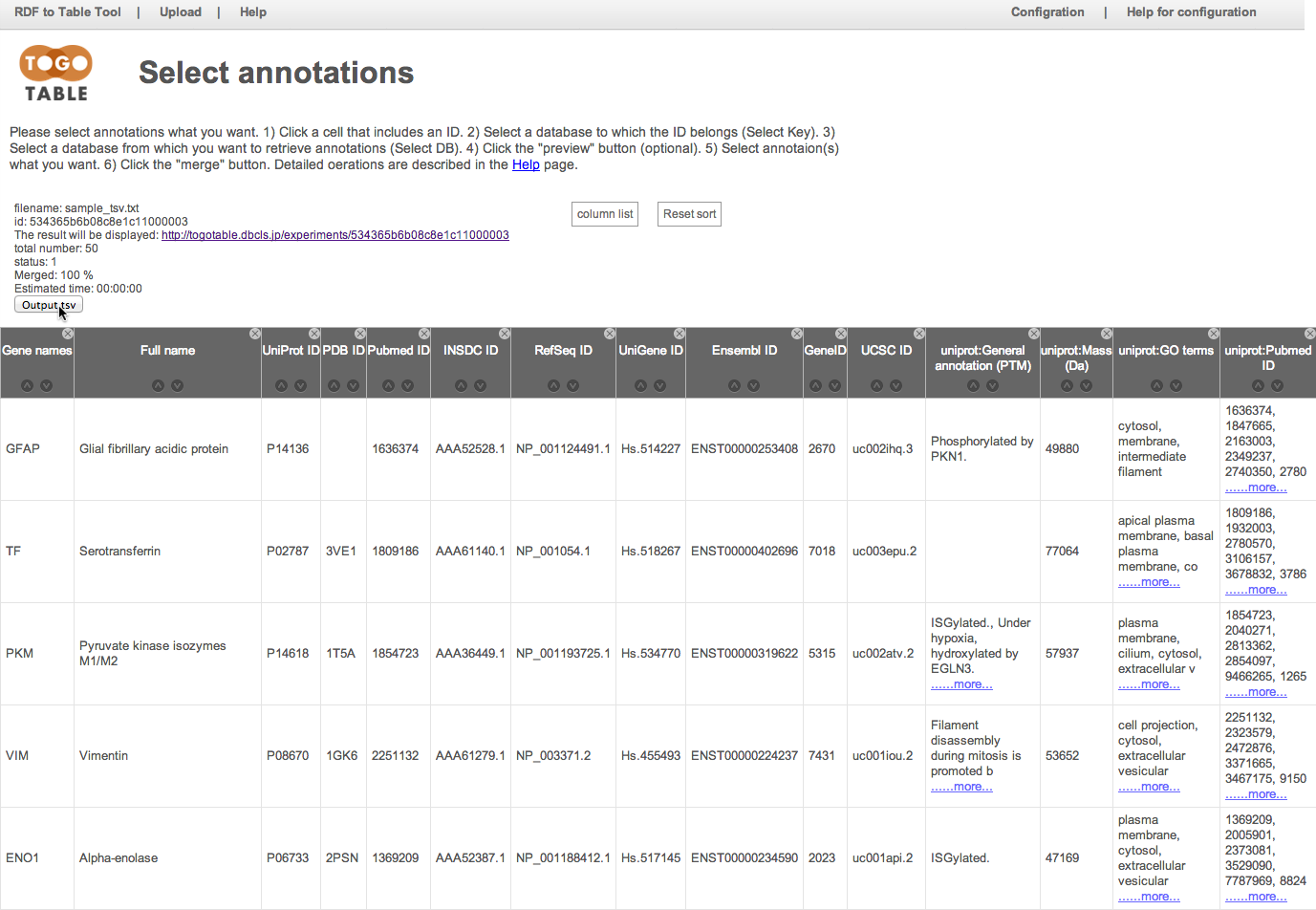
Click a cell that includes an ID
"P08670" in the UniProt ID column is selected here.
Currently, you can select UniProt ID, PDB ID, PubMed ID, INSDC (Genbank/ENA/DDBJ) ID, RefSeq ID, UniGene ID, Ensembl ID, GeneID, KEGG GENES ID, and UCSC ID.
Note that IDs are case-sensitive except for the PDB ID.
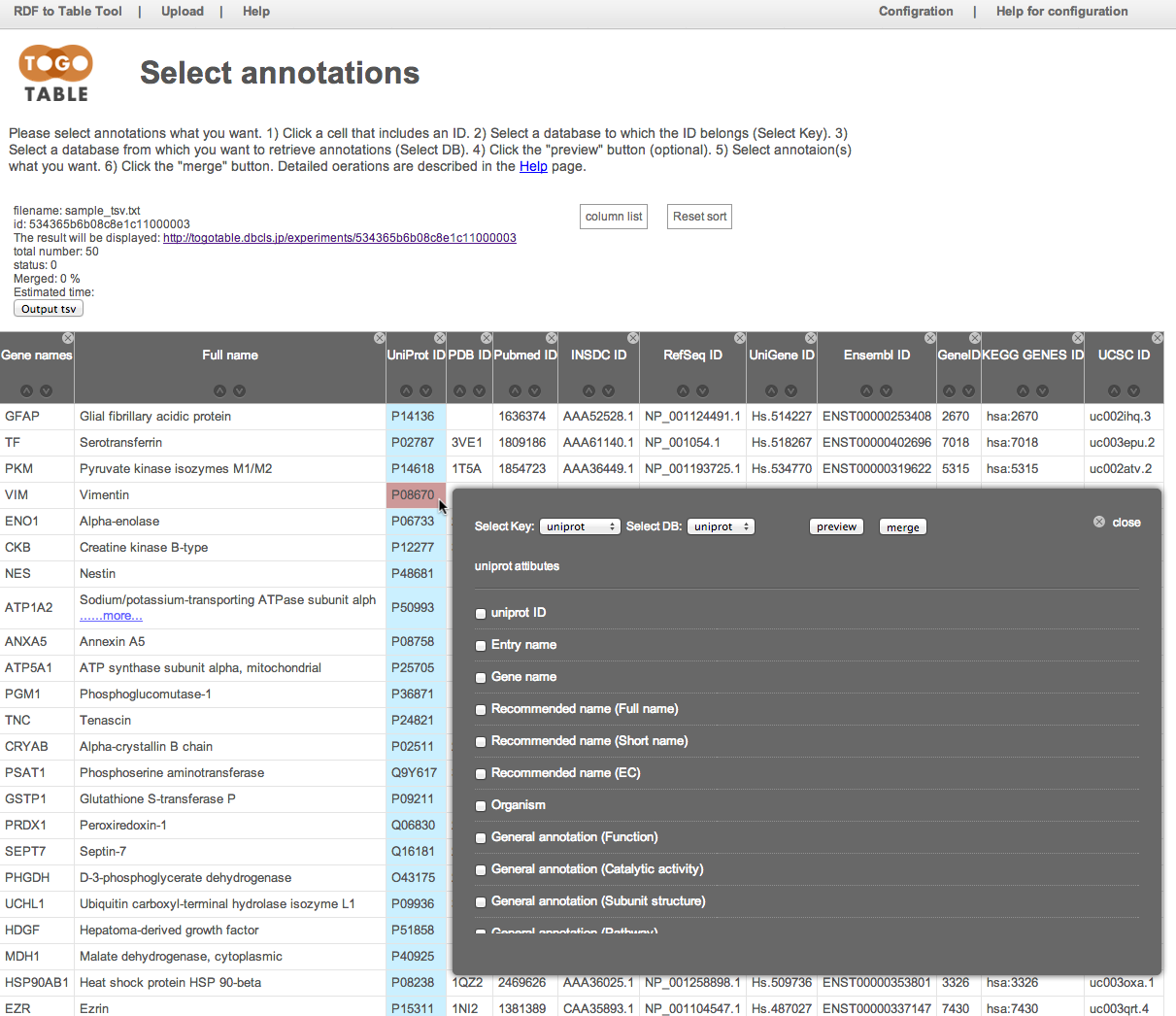
Select the database to which the ID belongs
Here, select "UniProt" from the database list in the "Select Key"
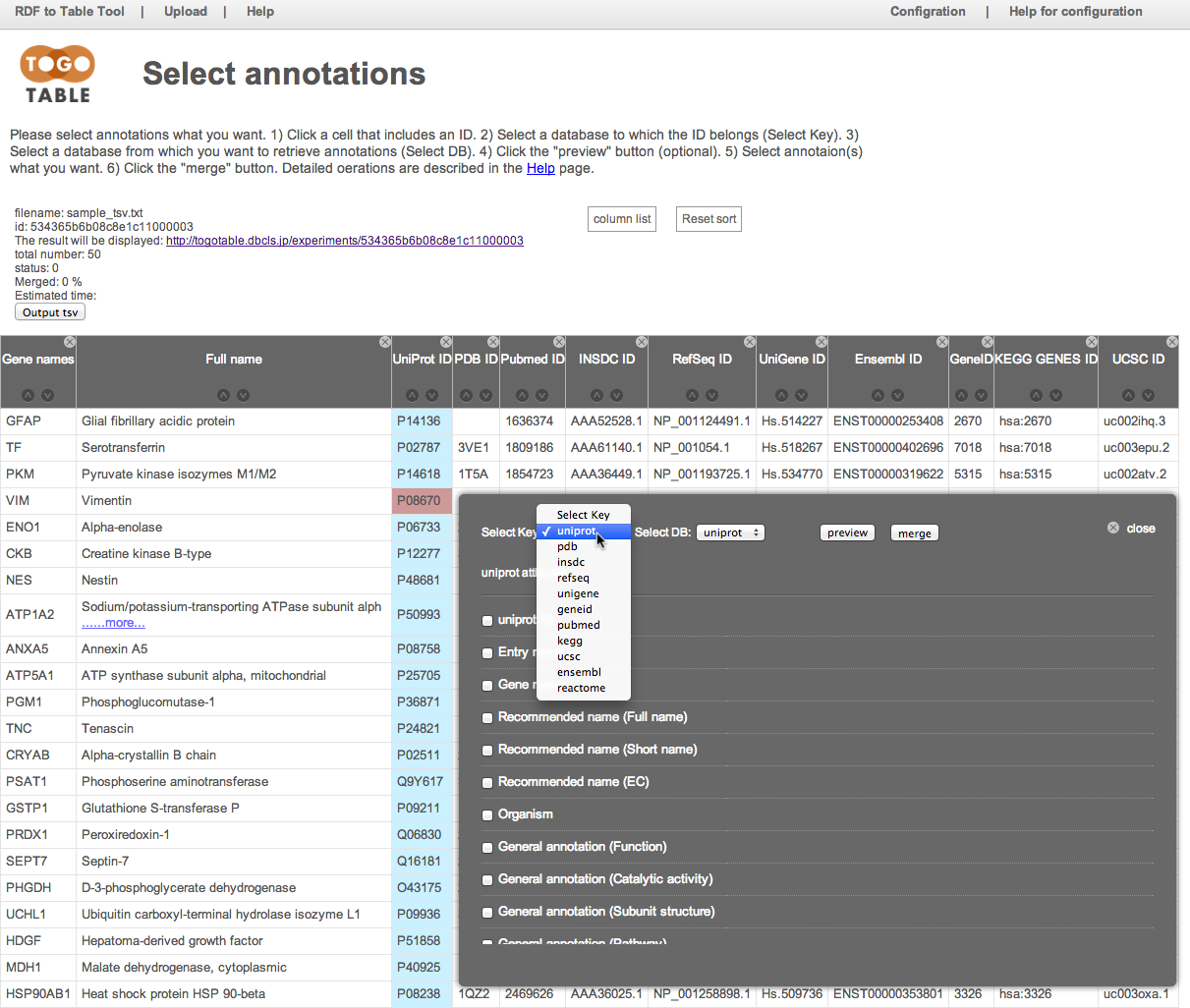
Select the database from which you want to retrieve annotations
Here, select "UniProt" from "Select DB"
Of course, you can select a different database from the selected database to which the selected ID belongs
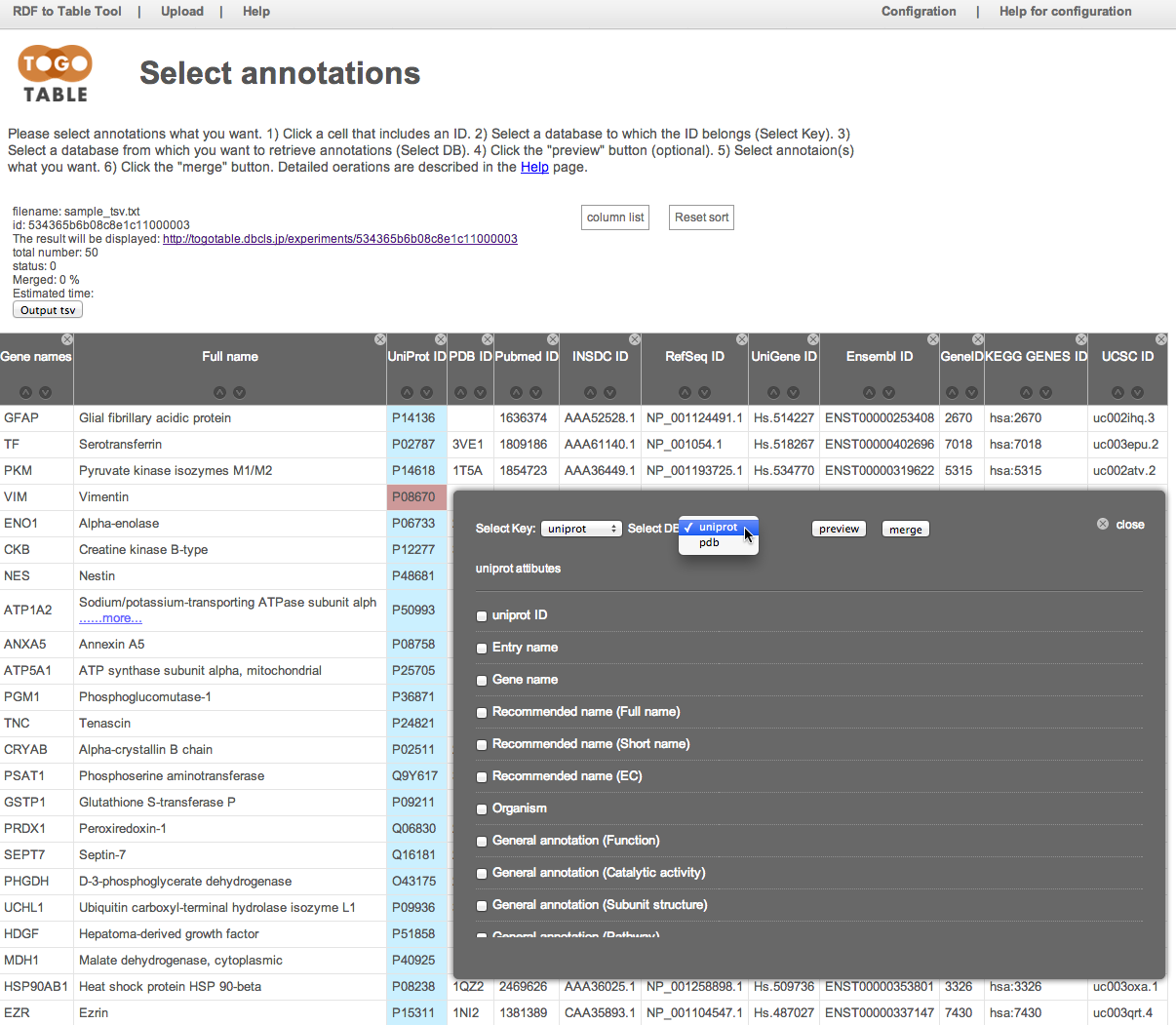
Select the annotation(s) that you want
Here, select "General annotation (PTM)", "GO terms", "Mass (Da)", and "PubMed ID"
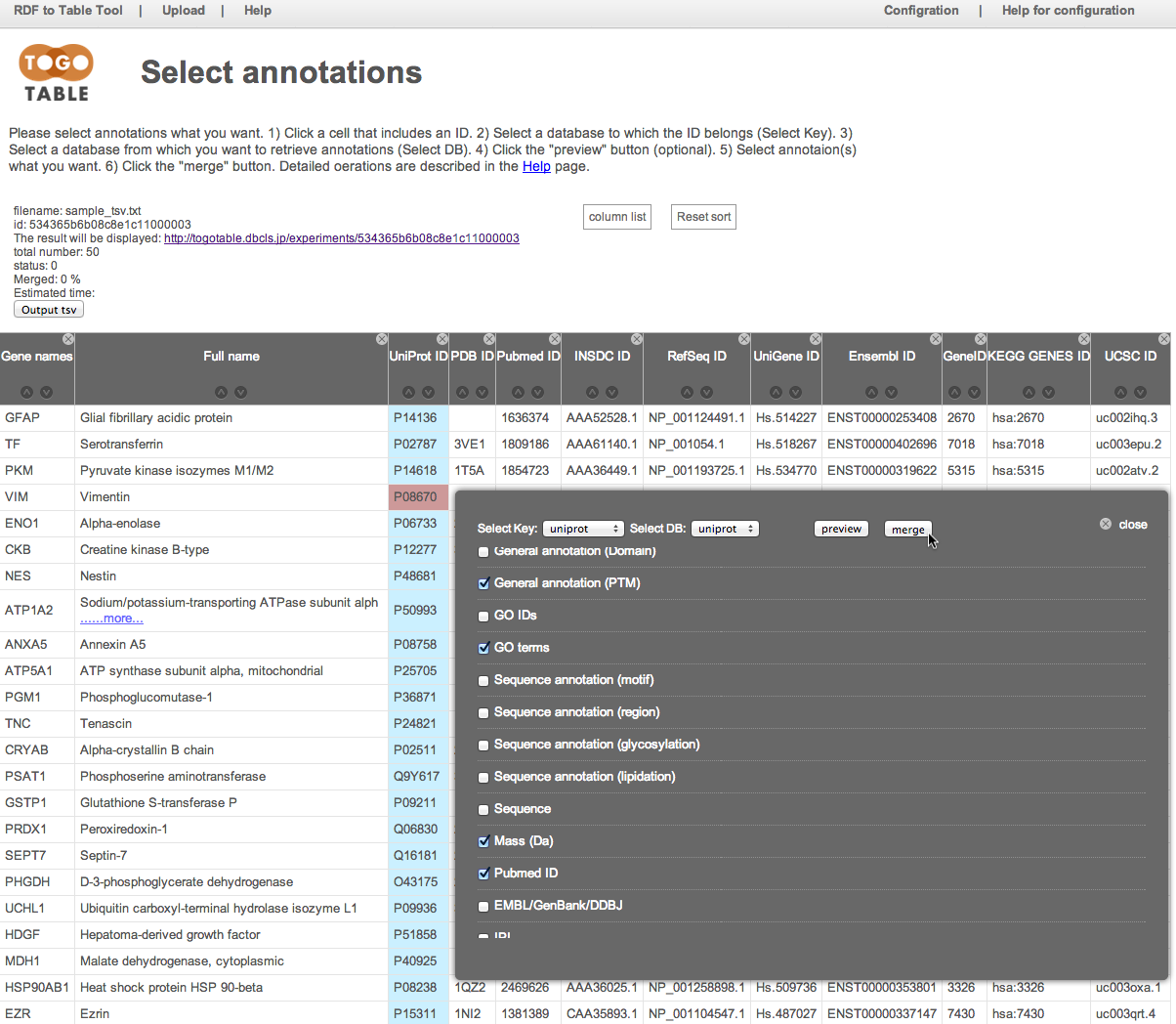
The selected annotations are displayed on the right side of the table (It may take time depending on the number of lines and the specified number of annotations)
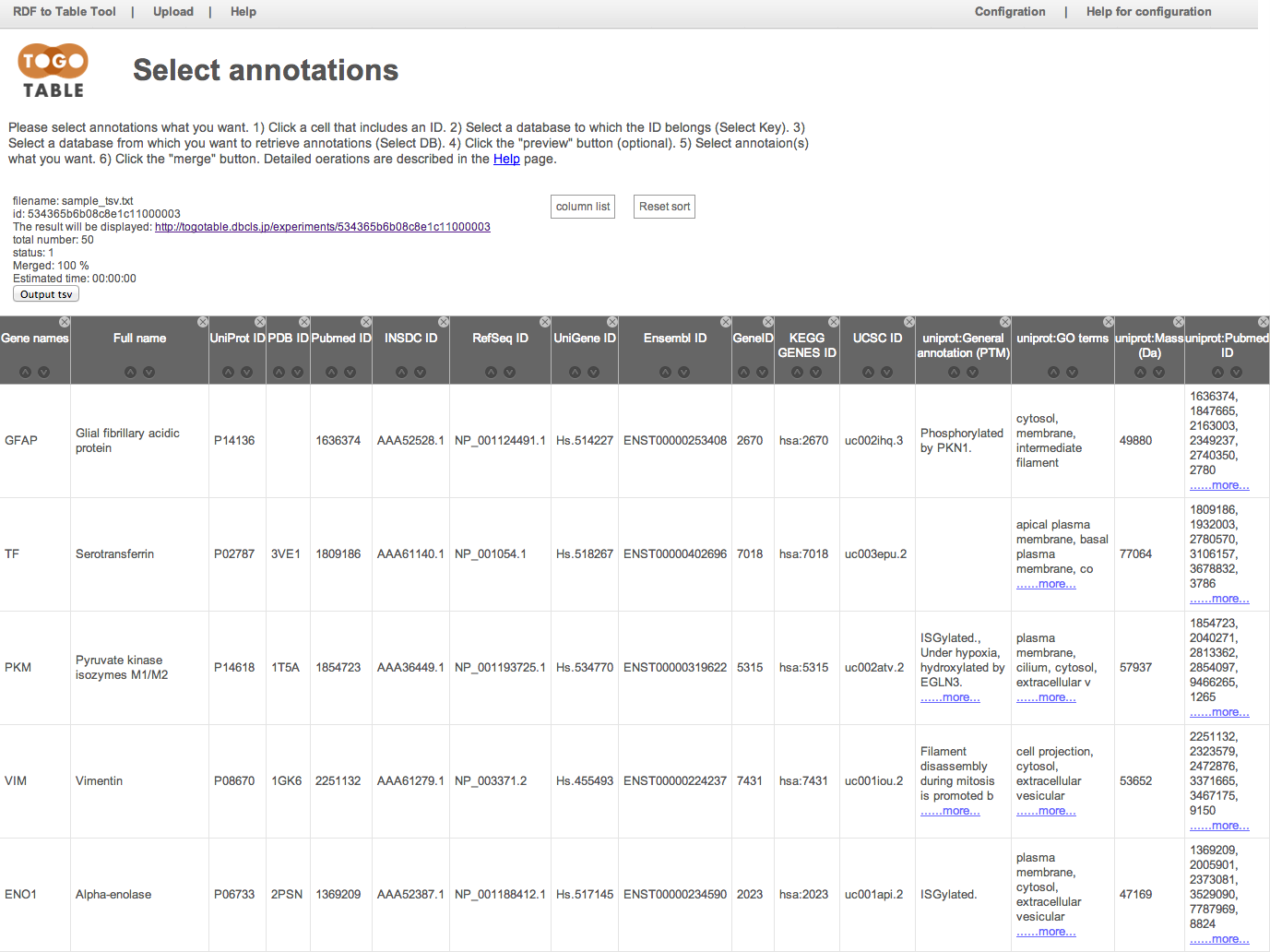
You can shuffle columns by dragging a column header
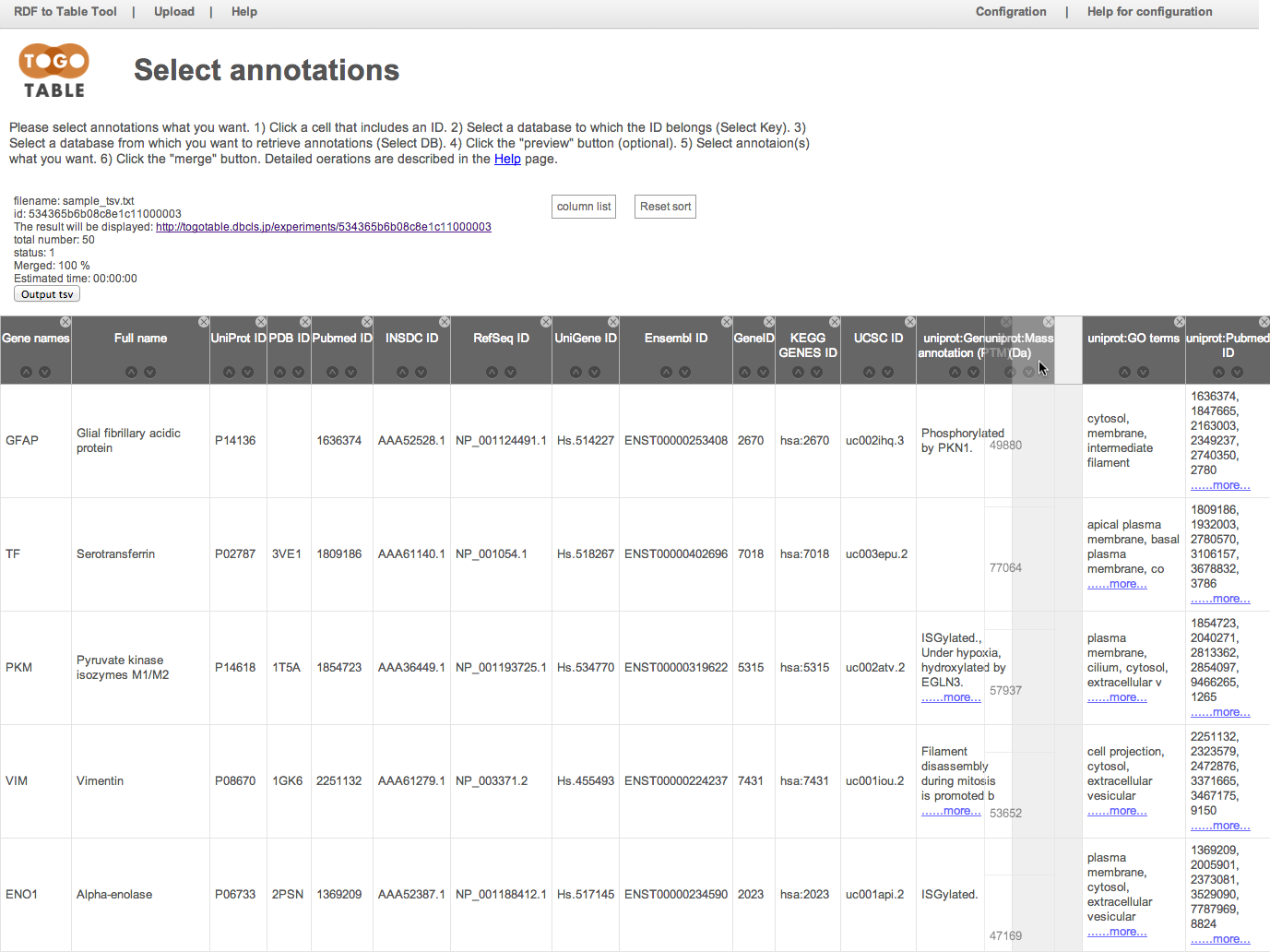
and you can hide a column by clicking a "x" mark on an upper right of column header
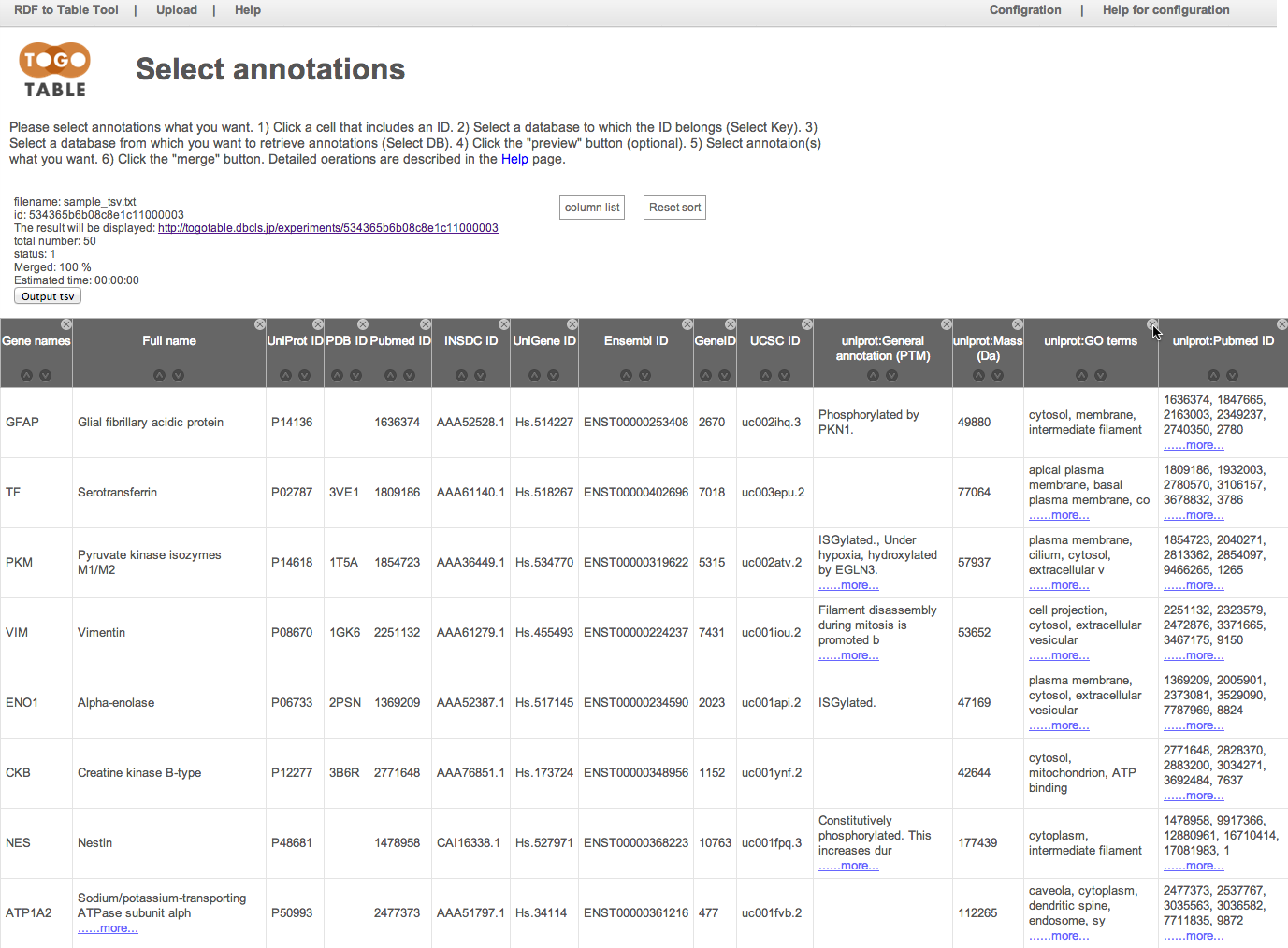
You can re-display the hidden columns by selecting checkboxes in the "column list"
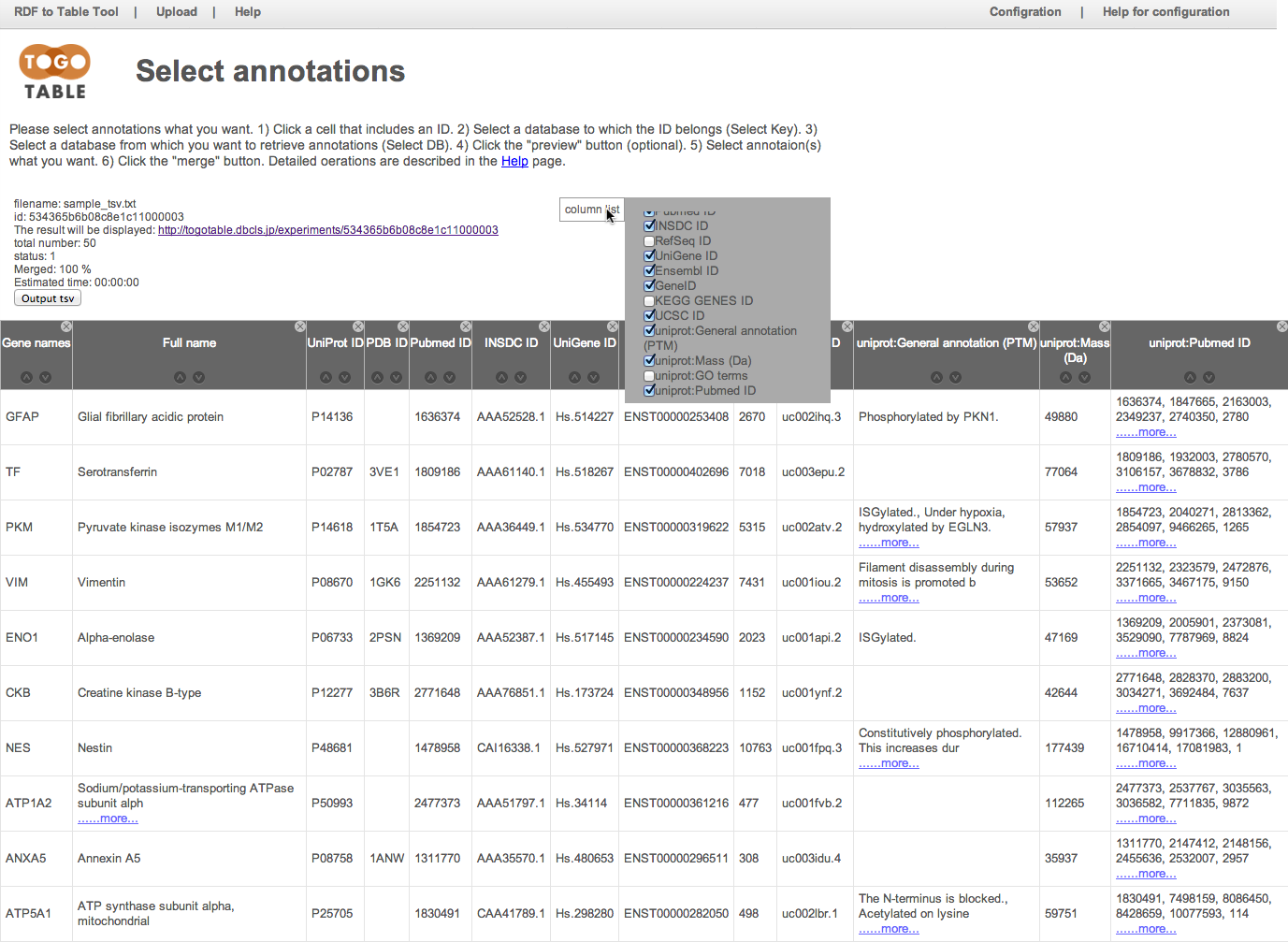
You can download the table as the TSV file, and it can be opened by standard spreadsheet applications