This page is a tutorial for adding new retrieveing annotations by users in TogoTable. Here, how to retrieve "pathway name" from reactome IDs in the Reactome database using its SPARQL endpoint is described as an example. The configuration is stored in the user's own browser.
How to use the TogoTable main system is here.
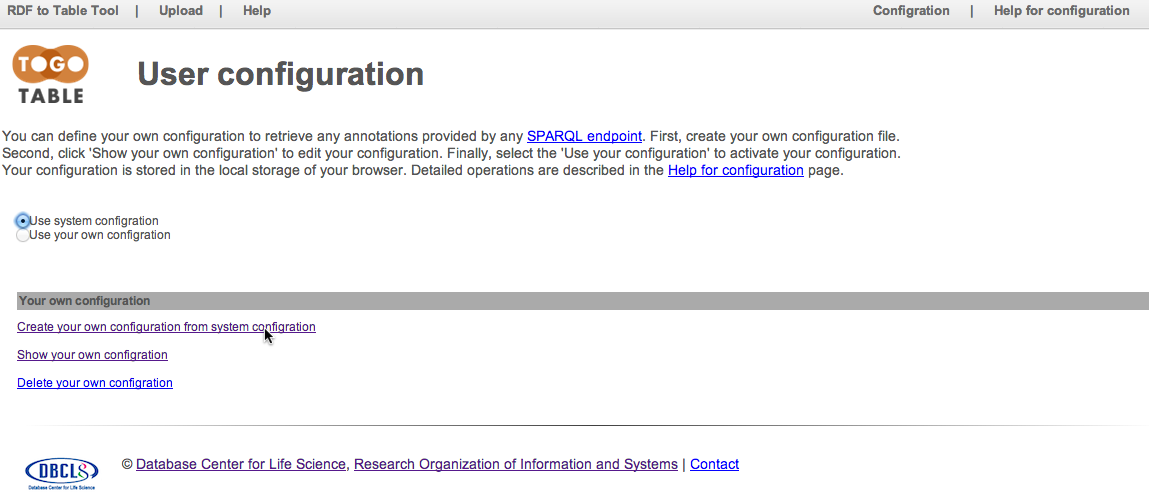
Create user's own configuration based on the system configuration
Click "Configuration" on the upper right of TogoTable.
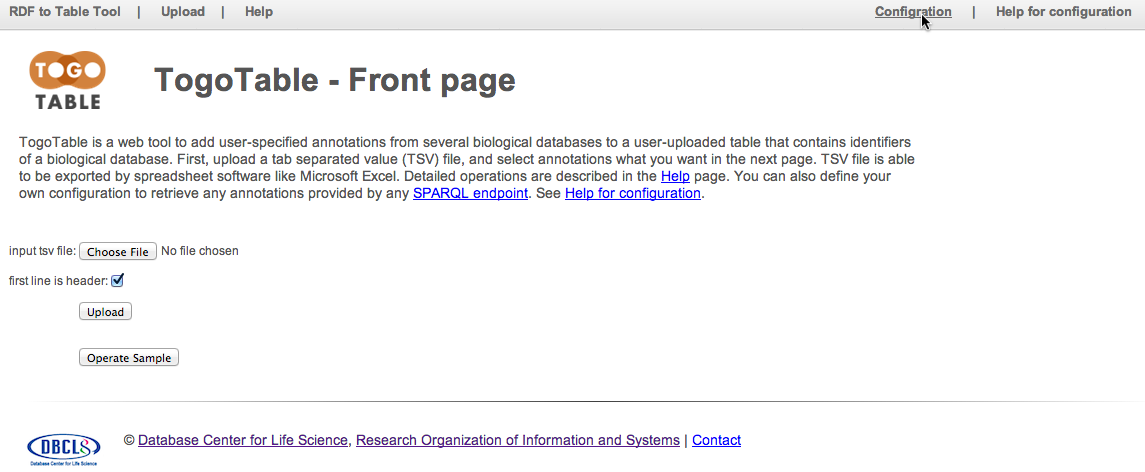
Click "Create your own configuration from system configuration."
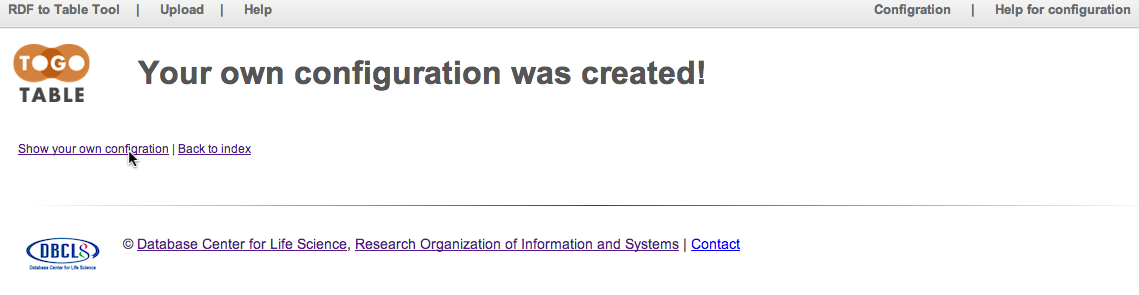
User's own configuration is created. Click "Show your own configuration."
Configure database to search (SPARQL endpoint)
Current registered databases are shown. Click "new DB"
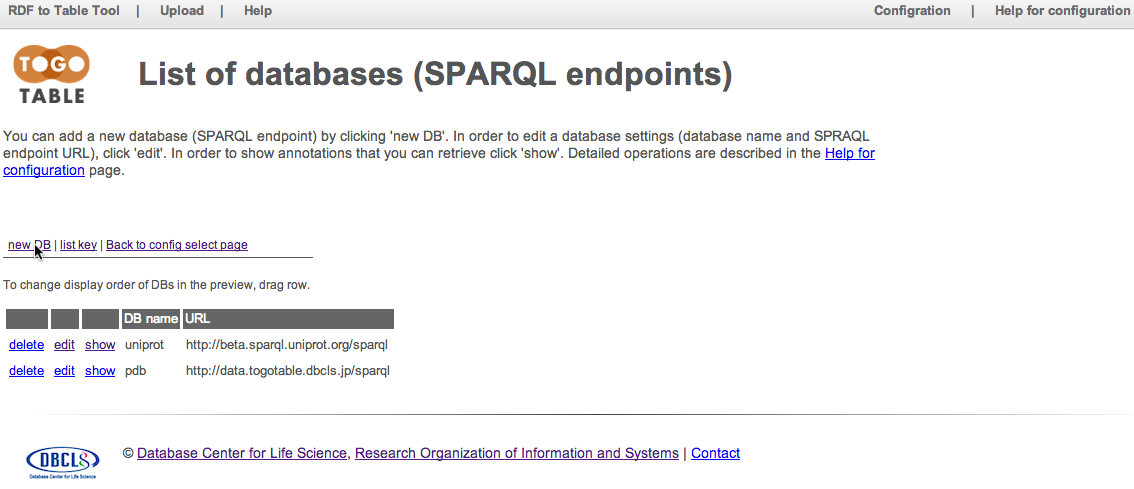
Specify a database name, SPARQL endpoint URL, subject URI prefix, and (if desired) a regular expression of IDs. Note that the database name should be unique in the configuration and should not include spaces. Here, set "reactome" to "DB name", "http://www.ebi.ac.uk/rdf/services/reactome/sparql" to "server URL", "http://identifiers.org/reactome/" to "subject prefix", and "REACT_[0-9]+\.[0-9]+" to "key regexp"
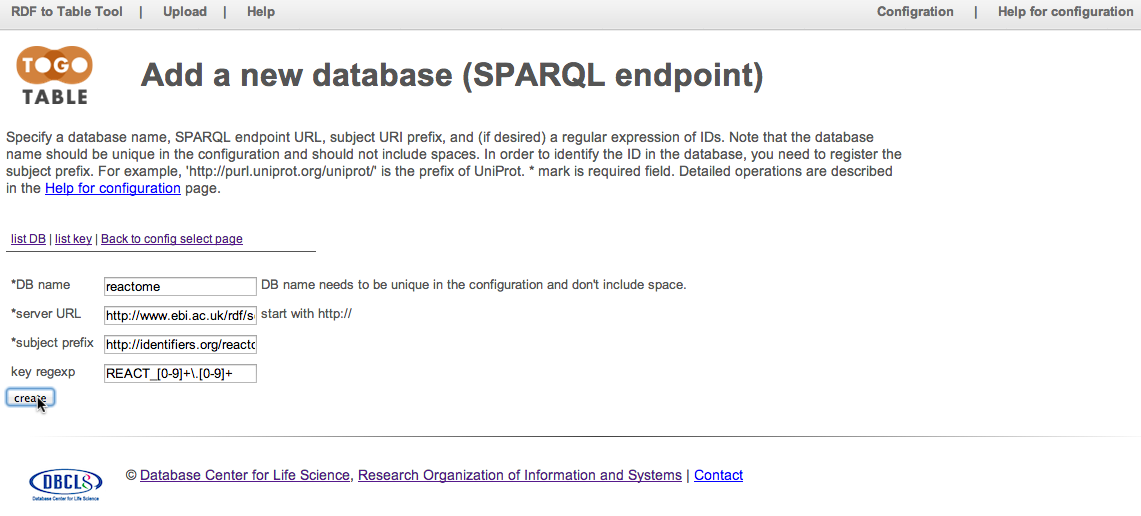
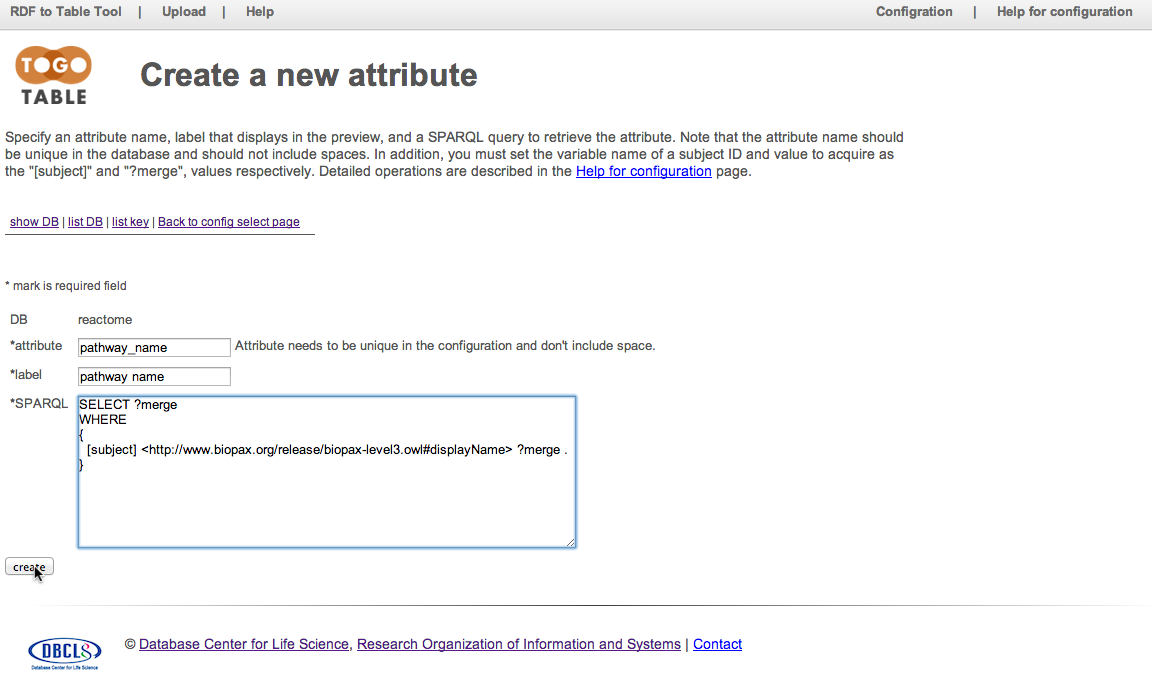
Reactome SPARQL endpoint is registered. Click "show" in the "reactome" row to configure attributes to retrieve.
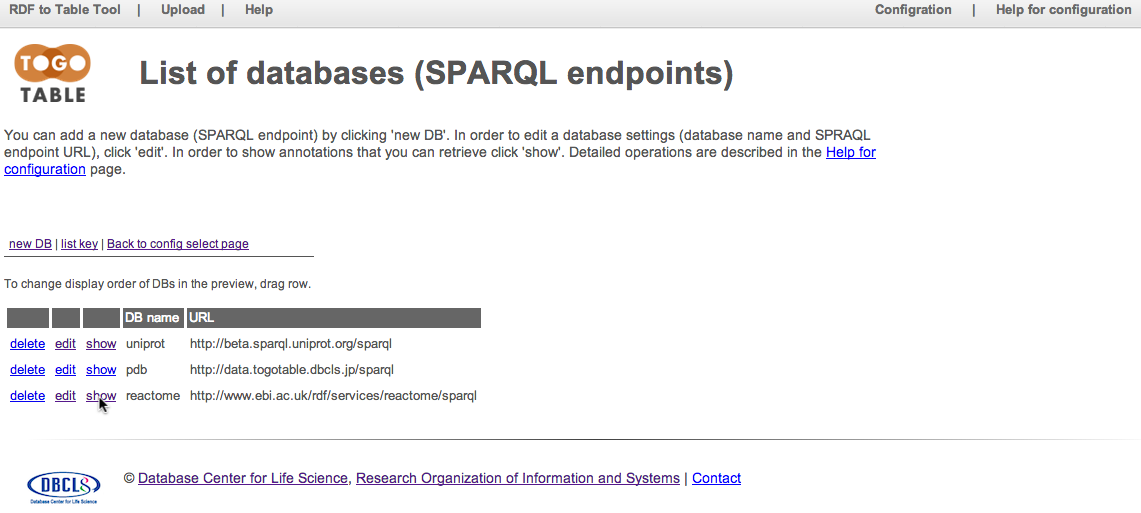
Configure an attribute to retrieve
Click "new attribute"
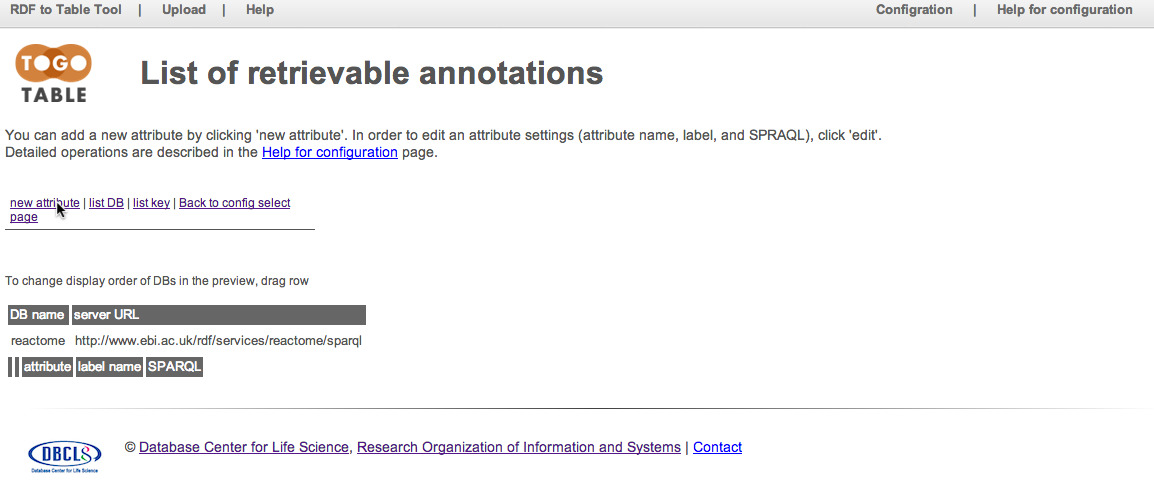
Specify an attribute name, label that displays in the preview, and a SPARQL query to retrieve the attribute. Note that the attribute name should be unique in the database and should not include spaces. In addition, you must set the variable name of a subject ID and value to acquire as the "[subject]" and "?merge", values respectively. Here, set "pathway_name" to "attribute", "pathway name" to "label", and the SPARQL command described below to "SPARQL"
SELECT ?merge
WHERE
{
[subject] <http://www.biopax.org/release/biopax-level3.owl#displayName> ?merge .
}
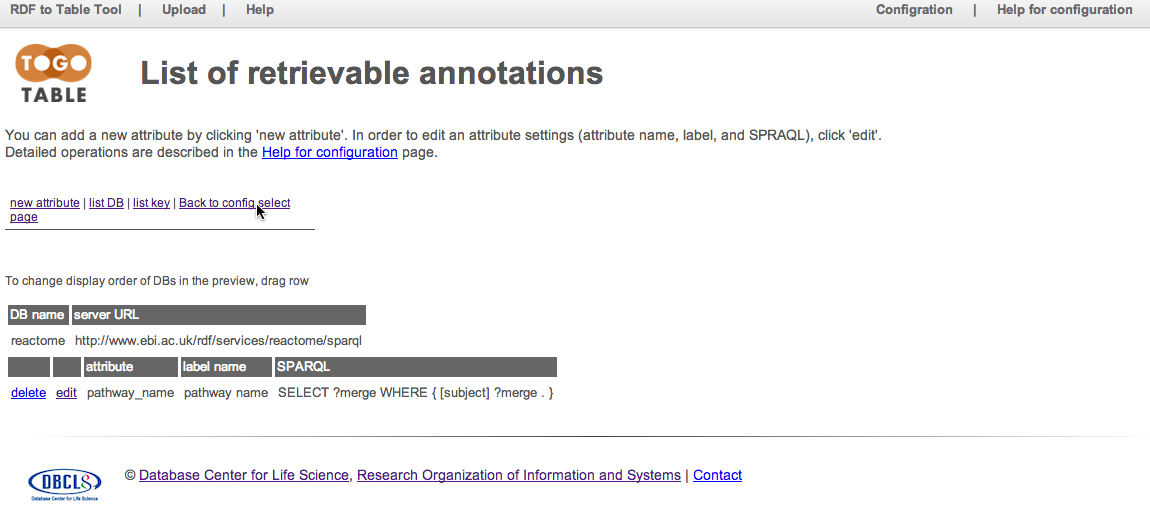
Configuration for retrieving pathway names from the Reactome database is saved. Click "Back to config select page" to return the top page of the configuration.
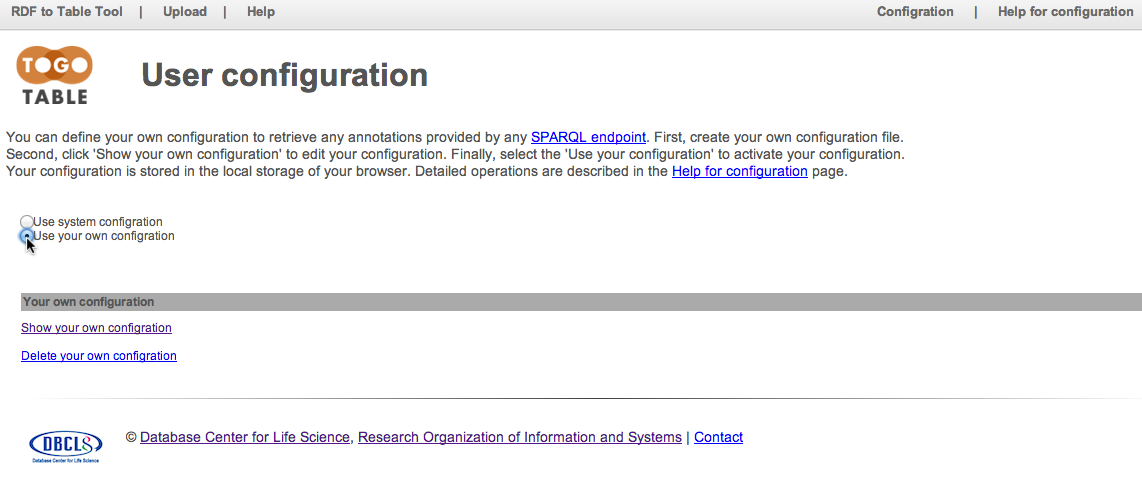
Select "Use your own configuration" to active uer's configuration in the TogoTable
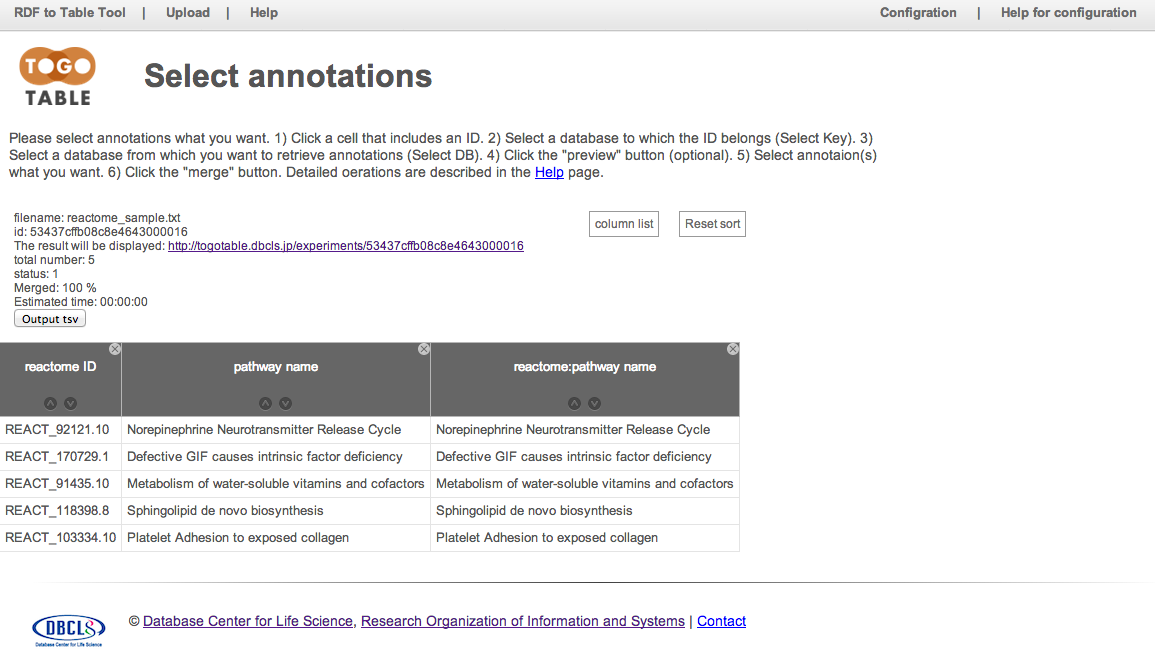
Retrieve pathway names from reactome IDs
Upload a file including Reactome IDs on the TogoTable top page. Click a cell including Reactome ID and select "reactome" in "Select Key" and "Select DB". Select the check box of "pathway name" and click "merge."
A sample file is here.
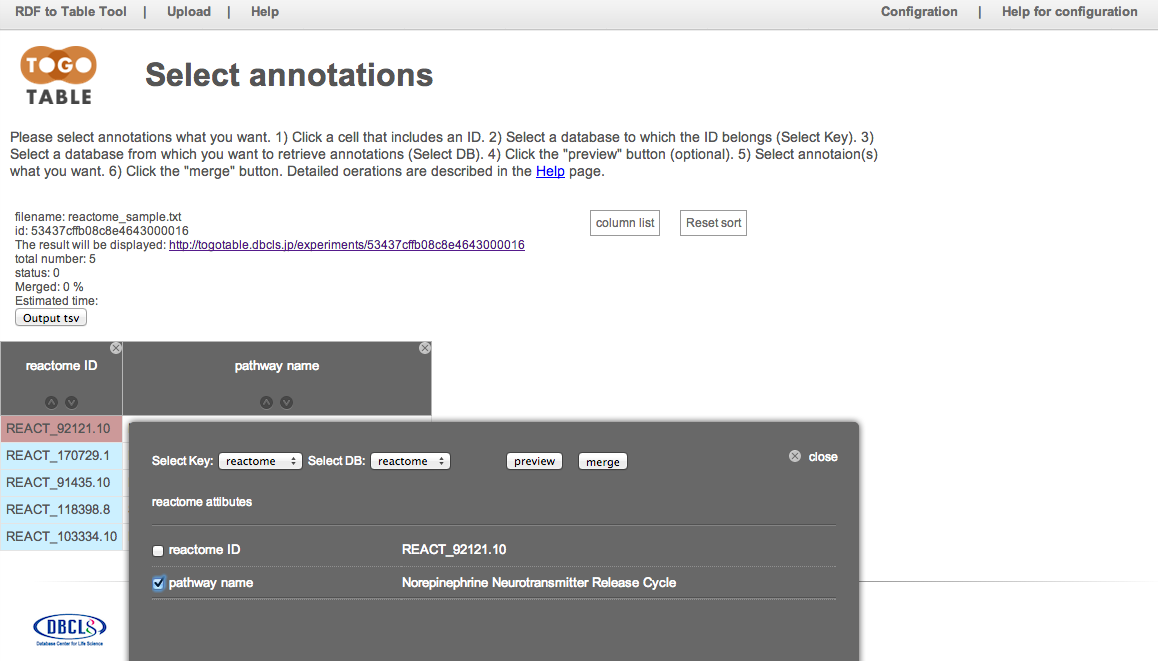
Pathway names are retrieved and displayed from the Reactome database via its SPARQL endpoint.